Utilization of Smut Resistant Primer (SRP) 3 in Evaluating the Resistant Hybrid Offspring from a Resistant and Susceptible Saccharum spp (Sugarcane) Parental Accessions
| Authors | JZ Louise P. Pantua, Hermie Ann F. Dela Peña, Trexie Pia C. Villarosa, John Moises G. Relles, Dynah Fatima I. Discaya, Meliza Z. Mana-a, Phillip Raymund R. De Oca |
|---|---|
| Volume | 1 |
| Date Published | September 27, 2021 |
| Date Update | September 21, 2024 |
Abstract
This study determines the resistance of Hybrid Sugarcane Offspring to Smut from pairs of resistant and susceptible parental accessions using Smut Resistance Primer 3 (SRP3). Starting from cross-pollination, followed by the extraction of 60 leaf samples using the CTAB method, then checking their concentration and purity. Agarose Gel Electrophoresis and PCR screening were done to check the quality, and amplification and copying of DNA segments. Polyacrylamide Gel Electrophoresis was performed to obtain results subjected to Molecularmarker scoring and Cluster analysis using R software. Results revealed that cluster 1 with 32 hybrid accessions tested was found to be resistant to the disease. Results also showed that a resistant accession used by the male parent would yield more resistant offspring. It is recommended that the experiment should involve field tests to ensure the checking of phenotypic expression with the results to be optimized and used to facilitate the sugarcane breeding process in the country.
Introduction
Sugarcane is primarily an industrial crop as the cane is supplied to sugar industries, contributing 140 million USD to the Philippines’ economy, making white sugar, jiggery (gur), and other by-products like bagasse and molasses[1]. Therefore, the industry must produce products with good quality sugarcane.
Negros Occidental, in the Central Philippines, has always been known as a sugar province, with half of its agricultural lowland being sugarcane plantation producing about 55% of the national production[1][2].
One of the factors affecting the yield of this province-wide crop is smut, a severe sugarcane disease caused by a fungus called Sporisorium scitamineum that usually has massive damage in tropical countries. It could cause losses up to 30 to 40% in plant crops, 70% in ratoons and could reduce 3-7% of the sucrose content in an infected cane [3][4][5][6][7][8].
To combat this disease, triazole fungicides are used as a dip treatment to protect seed-cane from infection, but it is only applied to mother stocks (nursery) since waste fungicide solution will be difficult to be disposed of[3][7][9][10]. Moreover, although the treatment works, side effects may harm the human body[3][11].
Another method used is hot water treatment, typically used in eliminating various diseases such as smut in seed canes since most infections are killed at a high temperature; lower than the lethal point for sugarcane[6][9][12][13]. However, there has been evidence that hot water treatment increases the susceptibility of the seed cane to succeeding infections and cane yield reduction[14][15].
Microsatellites or Simple Sequence Repeat (SSR) are essential in mapping genomes. A tool widely used for marker-assisted selections, plant genetics, and breeding programs[16][17][18]. It is a repetitive segment in the DNA scattered throughout Theme is usually made up of 2-5 base pairs, repeating 8-50 times[19].
This study made use of Smut resistant primer (SRP) 3 for its association to sugarcane smut resistance[20] . The primer is a Cyclic nucleotide-gated (CNG) channel. In definition, it is a nonselective cation channel first identified in retinal photoreceptors and olfactory sensory neurons, a single-stranded DNA sequence that defines which region of the DNA is amplified[21][22].
The goal of this study was to determine the resistance of the hybrid to the fungal disease. The findings would be added to the existing knowledge of the sugarcane genes’ resistance resulting from crossbreeding resistant and susceptible accessions.
Methodology
The study was performed at the Sugar Genome Laboratory, La Granja Agricultural Research and Extension Center (LGAREC), La Carlota City, Province of Negros Occidental, Philippines. LGAREC provided all chemical reagents and materials used for analyzing smut resistance.
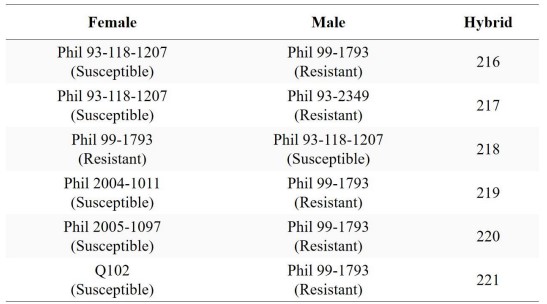
Table 1. Hybrids with their Parental Accessions
Choosing and Crossbreeding of the Parental Accessions
Parental accessions, one resistant and the other susceptible to smut disease, were chosen. Crossbreeding was done in a controlled environment where the female is placed in the soil while the male is placed above the female in a Hawaiian Solution. Every time the male plant pollinates, the pollen falls to the female.
Planting, growing, and retrieving of accessions from a resistant and susceptible parent gene
The seedlings were then planted in a seedbox located in a breeding nursery of SRA-La Granja and were grown for 3 months. The ten (10) leaf samples from six (6) different accessions were retrieved after 6 months.
Freeze drying and Extraction of the leaf samples using the CTAB Method
60 hybrid leaf samples were freeze-dried using ALPHA 1-2 LD plus Freeze Dryer for easier and efficient extraction. DNA was then extracted through the CTAB method. The CTAB Extraction Buffer and Polyvinylpyrrolidone are both effective in eliminating polysaccharides and polyphenols.
Identifying the Concentration and Purity of the extracted DNA samples
The concentration and purity of the extracted DNA samples were checked on the Nano-drop 8000 UV-Vis Spectrophotometer. The integrity or quality of DNA was checked through Agarose Gel Electrophoresis. The gel was made with calculated measurements, then the DNA samples were loaded on the wells; under the influence of constant current DNA fragments separated[23]. The Agarose gel was then exposed to UV light, the image of DNA showing its quality was viewed in the computer attached to the machine.
Making of the working solution
The working solution contains a calculated volume of gDNA, which is the V1 and C1, which is the concentration depending on the given results from the Nano-drop 8000 UV-Vis Spectrophotometer. There is also a fixed volume of 50µl of sNP (sterile nanopure) water and another C2 with a concentration of 100ng/µl
Polymerase Chain Reaction (PCR)
PCR mix was made containing a measured amount of 126µl Snp, 20µl PCR Buffer, 8µl MgCl₂, 4µl dNTPs, 10µl Primer_forward, 10µl Primer_ reverse, 2µl Taq Polymerase, 1µl gDNA. It was then added to the working solution using a pipette in a tube. All 57 samples were placed in the Bio-rad C1000 PCR machine for the stages of PCR screening. These three processes were repeated for 30x for DNA to be visualized using the Bio-rad C1000 PCR machine.
DNA Fingerprinting using Polyacrylamide Gel
Polyacrylamide Gel Electrophoresis (PAGE) was used to separate DNA fragments based on their size during this step. The gel was later viewed on the UV-Vis Spectrophotometer to view the bands that were formed.
Molecular Marker Scoring and Translating Data to Binary Data
After PAGE, molecular scoring was done to determine the number of bands present in each variety, the average allele for each primer, and the maximum and minimum size of the alleles. Evident and apparent bands were identified through Molecular Marker and were scored one in the presence and zero in the absence of DNA fragments.
Cluster Analysis
The installation of R-studio was done to encode the binary data that showed the Dendrogram results, which contains the dissimilarity of both susceptible and resistant hybrid accessions.
Recording, Analyzing, and Interpreting data and formulating a Conclusion
All data gathered from experimentation were collected, recorded, and encoded. The researchers were now able to analyze and interpret their data and formulate a concrete conclusion.
Results and Discussions
Purity of extracted leaf samples
The line graph below shows the purity of the extracted hybrid leaf samples. The average content of the purity level ranges from 1.8 to 2.0. If the purity of DNA is less than 1.8, it contains protein contamination. If the purity level is above 2.0, it possesses phenol contamination, and if the purity level ranges between 2.5-3.0, then polysaccharide was extracted instead of DNA. Based on the results, there is no polysaccharide extracted and phenol contamination, but there is a lot of protein contamination with 218-1 having the highest value of 0.19.
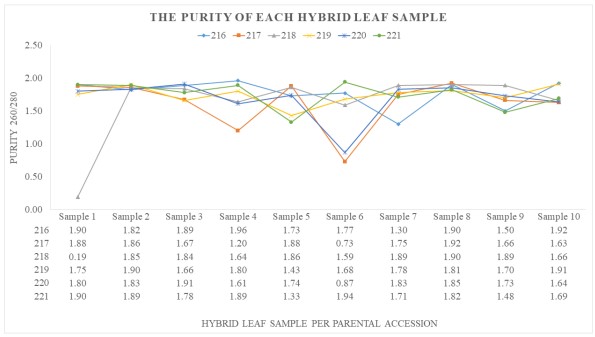
Figure 1. The purity of each extracted hybrid leaf sample
Polyacrylamide Gel Electrophoresis
There are 12 Primers in total. Each primer has a different number of alleles present. SRP 3-2 having the highest with 31 alleles; SRP 3-9 having 24; SRP 3-3 having 23 alleles; SRP 3-18 having 21 alleles; SRP 3-1 having 20 alleles; SRP 3-5 and SRP 3-7 having 18 alleles, and SRP 3-4 having 2 alleles. The average alleles per primer is 13.08. The maximum size of the bonds is 900 bp, while the lowest would be 100 bp.
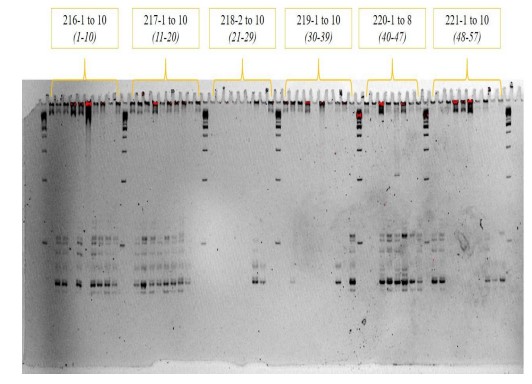
Figure 2. PAGE Results subjected to molecular marker scoring
Cluster Analysis
Figure 3 shows the dendrogram results through the R Statistical program. The statistical analysis was made with the help of two more expansive the R Statistical Language and Environment, wherein the Binary data was encoded to create the dendrogram. This provides an outline of the genetic distances of the accessions. The Diagram shows the hierarchical clustering of the relationships between similar sets of data. According to the results, there were 2 clusters shown, cluster 1 showing resistance and cluster 2 showing susceptibility to smut. Cluster 1 shows that 32 hybrid accessions are resistant, while Cluster 2 with 25 accessions is susceptible. These accessions have genetic markers that make them resistant to the disease. Dissimilarities show the genetic distances of the accessions based on the number of alleles detected. The dendrogram also contains the values of their dissimilarity. The closer the branch to 1.0, the more genetically distant the accession. Accessions with the highest dissimilarity of 1.0 are 216-1, 16-4, 216-6, 217-1, 217-10, 218-2, 218-3, 218-4, 218-5, 218-6, 218-7, 218-10, 219-3, 219-4, 219-5, 219-6, 219-7, 219-9, 220-1, 220-2, 221-3, 221-4, 221-5, 221-6, 221-7 and the lowest of 0.18 are 216-3 and 217- 3.
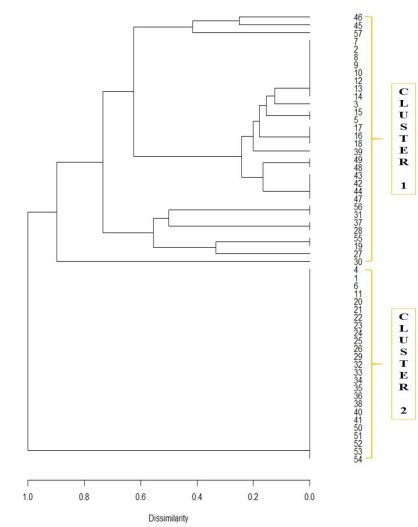
Figure 3. Dendrogram Results: 32 accessions (cluster 1), 25 accessions (cluster 2)
Conclusion
Based on the results, 32 hybrid accessions tested were resistant to sugarcane smut. 217, crossed from Phil 93-118-1207 (female, susceptible) and Phil 93-2349 (male, resistant) have 8 out of 10 of its samples tested resistant. 218, crossed from Phil 99-1793 (female, resistant), and Phil 93-118-1207 (male, susceptible) has the least, having only 2 out of 10 of its samples tested resistant. For a wider interpretation, the results were compared to the results of an umbrella project which made use of SRP 6 and 4 out of 32 resistant hybrid accessions came out to be resistant to smut as well. The following are: 43 which is 220-4, crossed from Phil 2005-1097 (female, susceptible) and Phil 99-1793 (male, resistant); 44 which is 220-5, crossed from Phil 2005-1097 (female, susceptible) and Phil 99-1793 (male, resistant); 56 which is 221-9, crossed from Q102 (female, susceptible) and Phil 99- 1793 (male, resistant); and lastly, 19 which is the 217-9, crossed from Phil 93-118-1207 (female, susceptible) and Phil 93-2349(male, resistant). Furthermore, when using the resistant accession Phil 99-1793 as the male parent, some offspring became resistant no matter which female parent was used. Another resistant accession used as the male parent, Phil 93-2349, showed similar results. Furthermore, 218, which used a resistant accession, Phil 99-1793, as the female parent resulted with the most susceptibility. Thus, utilizing a resistant accession as the father would more likely yield resistant offspring.
RECOMMENDATION
From the results, the researcher recommends growing the seedlings in the field, for they will be more exposed to natural conditions that enable the checking of their phenotypic expression. These accessions are hypothetically resistant. But for a more substantial result, they will have to undergo field testing.
References
- Moises, J., Relles, G., Laurena, A., Genaleen, M., Diaz, Q. & Lalusin, A. (2018). DEVELOPMENT OF MICROSATELLITE MARKERS FROM SUGARCANE (Saccharum Officinarum L.) PHIL 97-3933. International Journal of Agriculture, Environment and Bioresearch, 3(04).
- Liu, S., Lin, N., Chen, Y., Liang, Z., Liao, L., Lv, M., Chen, Y., Tang, Y., He, F., Chen, S., Zhou, J., & Zhang, L. (2017). Biocontrol of Sugarcane Smut Disease by Interference of Fungal Sexual Mating and Hyphal Growth Using a Bacterial Isolate. Frontiers in Microbiology, 8. https://doi.org/10.3389/fmicb.2017.00778
- Que, Y., Xu, L., Wu, Q., Liu, Y., Ling, H., Liu, Y., Zhang, Y., Guo, J., Su, Y., Chen, J., Wang, S. & Zhang, C. (2014). Genome sequencing of Sporisorium scitamineum provides insights into the pathogenic mechanisms of sugarcane smut. BMC Genomics, 15(1), 996.
- Id, E. (n.d.). TOPIC:WHIP SMUT OF SUGARCANE SUBJECT: BOTANY SEMESTER: 4 th SEMESTER M.Sc. BOTANY Paper/Course: MBOTEC-I (Unit V) (APPLIED MICROBIOLOGY AND PLANT PATHOLOGY).
- Sporisorium scitamineum (sugarcane smut). (2019). Cabi.org. https://www.cabi.org/isc/datasheet/55949
- Meena, B., & Ramyabharathi, S. A. (2012). Effect of fungicides and biocontrol agents in the management of sugarcane smut disease . Journal of Today’s Biological Sciences : Research & Review, 1(1), 96–103.
- A. Ramesh Sundar, E. Leonard Barnabas, P. Malathi & R. Viswanathan. (2012). A Mini-Review on Smut Disease of Sugarcane Caused by Sporisorium scitamineum. Intechopen.com. https://doi.org/10.5772/34116
- Shailbala, & Sharma, S.K.. (2013). Effect of fungicides and hot water treatment on control of sugarcane smut. 37. 29-32.
- Bhuiyan, S. A., Croft, B. J., & Tucker, G. R. (2015). New method of controlling sugarcane smut using flutriafol fungicide. Plant disease, 99(10), 1367-1373.
- Simmons, M. (2017, November 14). Triazole — toxicity, side effects,diseases, and environmental impacts.
- Bhuiyan, S. A., Croft, B. J., Deomano, E. C., James, R. S., & Stringer, J. K. (2013). Mechanism of resistance in Australian sugarcane parent clones to smut and the effect of hot water treatment. Crop and Pasture Science, 64(9), 892.
- Wondu, E. (2015). Design for Fabrication of Effective Seed Cane Hot Water Treatment Plant for Ethiopian Sugar Estates/Projects. International Journal of Science, Technology and Society, 3(5), 225.
- James, G. L. (1976, June). THE EFFECT OF RATOON STUNTING DISEASE ON THE EXPRESSION OF SMUT SYMPTOMS. Rhodesia Sugar Association Experiment Station, Chiredzi; Proceedings of The South African Sugar Technologists' Association.
- Efficacy of hot water treatment in management of sugarcane ratoon stunting disease | RUFORUM Institutional Repository. (2020). Ruforum.org. https://repository.ruforum.org/documents/ efficacy-hot-water-treatment-managemen t-sugarcane-ratoon-stunting-disease
- Varshney, R. K., Graner, A., & Sorrells, M. E. (2005). Genic microsatellite markers in plants: features and applications. Trends in Biotechnology, 23(1), 48–55.
- Hameed, U., Pan, Y.-B. ., Muhammad, K., Afghan, S., & Iqbal, J. (2012). Use of simple sequence repeat markers for DNA fingerprinting and diversity analysis of sugarcane (Saccharum spp) cultivars resistant and susceptible to redrot. Genetics and Molecular Research, 11(2), 1195–1204.
- Cordeiro, G. M., Taylor, G. O., & Henry, R. J. (2000). Characterisation of microsatellite markers from sugarcane (Saccharum sp.), a highly polyploid species. Plant Science, 155(2), 161–168.
- Abdurakhmonov, I. Y. (2016). Introduction to Microsatellites: Basics, Trends and Highlights. Microsatellite Markers. https://doi.org/10.5772/66446
- Relles, J. M. G., Discaya, D. F. I., Mana-Ay, M. Z., Meneses, N. S., Entima, R. G., & Armones, R. T. Association Analysis of Developed Simple Sequence Repeat (SSR) Markers to Fungal Disease Resistance.
- Mazzolini, M., Arcangeletti, M., Marchesi, A., Napolitano, L. M. R., Grosa, D., Maity, S., Anselmi, C., & Torre, V. (2018). The gating mechanism in cyclic nucleotide-gated ion channels. Scientific Reports, 8(1). https://doi.org/10.1038/s41598-017-1849 9-0
- Garibyan, L., & Avashia, N. (2013). Polymerase Chain Reaction. Journal of Investigative Dermatology, 133(3), 1–4.
- kazilek. (2010, April 2). Electrophoresis | Ask A Biologist. Asu.edu. https://askabiologist.asu.edu/agarose-gelelectrophoresi