Smut Disease Susceptibility of Hybrid Offsprings from Resistant and Susceptible Saccharum spp (Sugarcane) Parental Accessions using Smut Resistant Primer (SRP) 6
| Authors | Julianne Marc M. Tamayo, Tisha Gabrielle P. Hormigos, Lyanne Clich S. Supe, John Moises G. Relles, Dynah Fatima I. Discaya, Meliza Z. Mana-ay, Phillip Raymund R. De Oca |
|---|---|
| Volume | 1 |
| Date published | September 27, 2021 |
| Date updated | September 21, 2024 |
Abstract
Smut disease susceptibility of different crossbred offsprings from resistant and susceptible sugarcane accessions were determined using Smut Resistant Primer 6 (SRP6). DNA samples were extracted from each hybrid accession. Their concentration, purity, and qualities were determined. Screening used PCR, then Polyacrylamide Gel Electrophoresis with the results subjected to Molecular Marker Scoring and Cluster Analysis using R Software. The results revealed that 42 offsprings were proven susceptible to Smut. When accession Phil 99-1793, resistant, is used as a male parent, some of the hybrids become susceptible, no matter which accession it is paired with. When compared to another primer with the same offsprings, accession 219 showed the most susceptible samples, while 217 showed none. The library generated by the marker scoring and R analysis from the study can be optimized to facilitate breeding in the country.
Introduction
Sugarcane farms mostly encounter diseases caused by either biotic or abiotic factors [1][2] . One of the most severe is Smut, a sugarcane fungal disease brought by the fungus Sporisorium scitamineum [3][4][5][6] . The disease can cause heavy losses to cane production, sugar content, and juice quality [7]8][9] . Canes that are infected indicate substantial metabolic changes leading to the formation of a whip-like sorus made up of a combination of fungal hyphae and plant tissues [10] .
The said disease hinders the target output of the Sugarcane Regulatory Administration anchored on the Sugarcane Industry Roadmap 2020, including: an increase in the sugarcane area, farm productivity, and sugar yield [11].
To safeguard stalk cuttings from the disease, triazole fungicides have been widely employed, in the likes of propiconazole and triadimefon [3][6][12] . The treatment, however, may be difficult to dispose of in large volumes and has logistics limitations as well as side effects on the human body [13][14] .
Simple sequence repeats (SSRs), also called microsatellites, are proven and useful markers frequently utilized as a resourceful tool in plant breeding programs and evolutionary studies due to their significant ability in indicating cultivar diversity [15][16][17][18] .
A microsatellite is a span of repetitive genetic sequence in which the repeating unit contains from one to six bases [19] . Microsatellites are used to locate a gene or a mutation responsible for a given trait or disease. They can be amplified for identification by PCR and can be used as molecular markers [20] . Through microsatellites, the sugarcane varieties being released are screened and tested through their genes or DNA.
The Smut Resistant Primer used for significant association with susceptibility in this study is SRP6 [2] . This primer is a serine/threonine kinase protein. This protein plays a paramount role in cellular homeostasis [21] . Short fragments of DNA with a specific sequence corresponding to the target DNA to be identified and amplified are the primers [22] . In this study, susceptibility to the Smut disease of different accessions of sugarcane that are offsprings of recessive and susceptible crossbred accessions was determined. This study seeks to help in the development of markers to quicken the release of varieties where the selection usually spans a period of 12 years [23] , thus increasing the yield and quality of sugarcane in the industry.
Methodology
Hybrid sugarcane offsprings crossbred from the following accessions were evaluated for susceptibility to the Smut disease with SRP6: Hybrid 216 is the offspring of Phil 93-118-1207 (female, susceptible) and Phil 99-1793 (male, resistant), 217 of Phil 93-118-1207 (female, susceptible) and Phil 93-2349 (male, resistant), 218 of Phil 99-1793 (female, resistant) and Phil 93-118-1207 (male,susceptible), 219 of Phil 2004-1011 (female, susceptible) and Phil 99-1793 (male, resistant), 220 of Phil 2005-1097 (female, susceptible) and Phil 99-1793 (male, resistant), and 221 of Q102 (female, susceptible) and Phil 99-1793 (male, resistant). This was conducted in the Sugar Genome Laboratory La Granja Agricultural Research and Extension Center.
Preparation of Crossbreed Accessions
The parental accessions were chosen wherein one is resistant and the other is susceptible. Cross-pollination was employed where the male is positioned above the female and placed in a Hawaiian solution: a mixture of acids formulated to give life to sugarcane every time it provides pollen. For the pollen of the male sugarcane to fall directly to the female every time it pollinates, the female sugarcane is placed in the soil below the male sugarcane. The aim is to identify the gene sequence responsible for the sugarcane variety’s susceptibility to Smut. The seedlings were grown in a greenhouse nursery.
Extraction of DNA Samples
After six months, ten (10) samples each from six crosses were obtained. The samples were freeze-dried with ALPHA 1-2 LDplus Freeze Dryer. The process is done for extraction efficiency and total removal of moisture from the leaves. Extraction of the freeze-dried samples used the CTAB method, eliminating contaminants, polysaccharides, and polyphenols by applying the cationic detergent CTAB and Polyvinylpyrrolidone: the polyphenol binding agent.
Evaluation of the Concentration and Purity of the Samples
NanoDrop 8000 Ultraviolet-Visible Spectrophotometer was used to check the concentration and purity of the extracted samples. The quality of the extracted samples was checked through Agarose Gel Electrophoresis. It followed three protocols: first, a gel was prepared with an agarose concentration adjusted to match the size of the DNA fragments to be separated. Sample wells were loaded with the DNA samples. The gel was run at 100V for a period of time until optimal separation was attained. Finally, using a Bio-Rad Gel Doc XR+, the gel was visualized directly after being illuminated with UV light [23] .
Preparation of Working Solutions
The formula C1V1=C2V2 was used to prepare volume 1 of the working solution, wherein C stands for concentration and V for volume. Volume 1 is the volume of the sample to be taken from the stock solution. The calculated amount of Volume 1 was added individually in separate tubes and added sNP (sterile nanopure) water to each sample.
Polymerase Chain Reaction
PCR mix: SnP water, ICR buffer, dNTPs, MgCl2, Primer forward, Primer reverse, and Taq Polymerase were prepared. The reagents were pipetted in a 1 mL tube in order previously stated. The PCR mix prepared for the initial working solutions were added. The stages of the PCR Screening were Denaturation (95°C), Annealing (51°C), Elongation (72°C), and 12°C. The cycle was repeated 30 times for DNA to be visualized
Polyacrylamide Gel Electrophoresis and Gel Documentation
Polyacrylamide gel electrophoresis (PAGE) was used. In this, for binding to proteins and giving them a negative charge, Sodium dodecyl sulfate (SDS) was utilized. Then, the proteins were electrophoretically sorted by size through a gel matrix composed of polyacrylamide in an electric field. Bio-Rad GelDoc XR Plus was then used for the Gel Documentation Process.
Molecular Marker Scoring and Cluster Analysis
Each of the PAGE run was subjected to molecular marker scoring. Only evident and apparent bands were scored following the procedures of Relles et al. Bands were scored one (1) for the presence and zero (0) for the absence of a DNA fragment. The presence or absence of the amplified band in all genotypes indicates similarity, whereas the presence on one and absence on another indicates dissimilarity[2] . The software R studio was used where the binary data were encoded and the dendrogram was generated.
Results and Discussions
Purity of the Extracted Samples
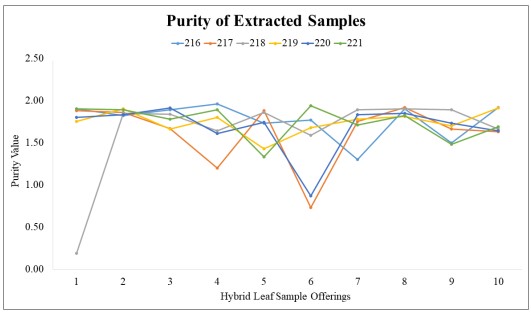
Figure 1. Initial 60 extracted samples’ purity
Figure 1 shows the results for the purity of the 60 extracted samples. The purity level values range from 1.8 to 2.0. When the purity level hits lower than the said range value, it means that the DNA extracted contains protein contamination. If the purity level is above 2.0, the DNA extracted contains phenol contamination. When the purity level reaches 2.5-3.0, then the polysaccharide was extracted instead of DNA in the hybrid leaf sample. The sample with the highest purity value based upon the figure is 220-4. The sample with the lowest is 218-1, with a value of 0.
Polyacrylamide Gel Electrophoresis
Figure 2. PAGE Results subjected to molecular marker scoring
Figure 3. PAGE Results subjected to molecular marker scoring
Based on the results from the Polyacrylamide Gel Electrophoresis, the bands are scored according to their positions in the ladder. The bands that can be visualized are those apparent only. They were counted and scored to know which variety has the highest amount of bands. The hybrid offsprings were labeled one (1) with the presence of bands and zero (0) when the bands are non-existent. Based on the results, 15 hybrid offsprings have bands, and the other 42 have zero (0) bands. Hybrid offsprings 218-2 and 218-6 have the highest number of alleles, which is 5. Hybrid offspring 218-5 and 221-5 have 4 alleles; hybrid offspring 218-7, 220-4, and 220-5 have 3 alleles; hybrid offspring 217-9, 219-7, 221-1, and 221-4 have alleles; and hybrid offspring 217-8, 221-7, 221-9, and 221-10 have 1 allele; and the other 42 hybrid offsprings have none. The average number of alleles per hybrid offspring is 0.64 alleles. SRP6-1 has 5 alleles, SRP6-2 has the most alleles detected with 14 alleles, SRP6-3 has 3, SRP6-4 has 8, SRP6-7 has 3, SRP6-8 has 1, SRP6-9 has 3, and SRP6-10 has 3. The primers with the lowest number of alleles present are SRP6-5 and SRP6-6 with 0 alleles. The maximum size of the alleles detected is 180bp, and the minimum is 130bp.
Cluster Analysis Using R Software
The dendrogram is a result of using the R Statistical Program. It is a visual representation that provides an overview of the genetic distances among accessions. Statistical analysis was done with the aid of the R Statistical Language and Environment. Encoding of binary data utilized this software in which it was interpreted to create the dendrogram. Cluster 1 shows the 15 offsprings that do not have a susceptible trait. Cluster 2 shows the 42 crossbred offsprings that have the probability of being susceptible. These offsprings have genetic markers in their genes that make them susceptible to the disease Smut. The Dissimilarity in the dendrogram signifies how genetically distant they are based on the alleles detected. The values below the dendrogram show the percentage of their dissimilarity. The closer the branch is to 1.0 (100%), the more genetically distant it is from the other accessions.
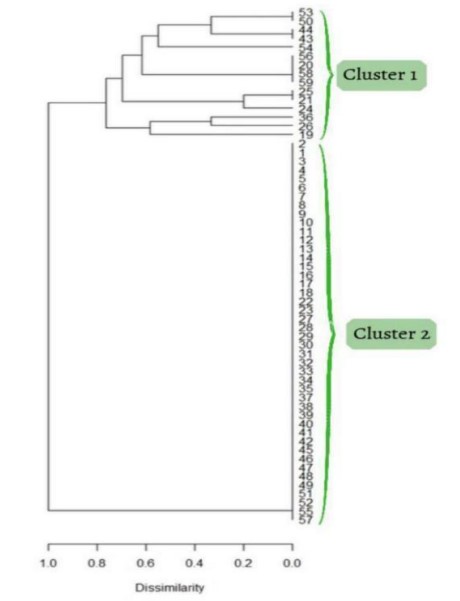
Figure 4. Dendrogram of the hybrid varieties showing Cluster 1 (15 varieties) and Cluster 2 (42 varieties)
The offsprings with the lowest dissimilarity is 218-4 and 218-5, garnering 0.21 (21%). This means that their genetic distance is close to each other. The offsprings with the highest dissimilarity of 1.0 (100%) are 216-, 216-2, 216-3, 216-4, 216-5, 216-6, 216-7, 216-8, 216-9, 216-10, 217-1, 217-2, 217-3, 217-4, 217-5, 217-6, 217-8, 218-2, 218-3, 218-7, 218-8, 218-9, 219-1, 219-2, 219-3, 219-4, 219-5, 219-6, 219-8, and 219-9, 219-10, 220-1, 220-2, 220-3, 220--6, 220-7, 220-8, 220-10, 221-1, 221-4, 221-6. They are 100% genetically distant from each other.
Conclusion
It was confirmed after tests that 42 crossbred offsprings had been proven susceptible to Sugarcane Smut disease with SRP6 as the primer used. Offspring 216, the cross between Phil 93-118-1207 (female, susceptible) and Phil 99-1793 (male, resistant) was shown to have the most susceptible samples. All ten samples of the offspring were found to be susceptible. 221, on the other hand, the cross between Q102 (female, susceptible) and Phil 99-1793 (male, resistant) was shown to have the least susceptible samples, with a total value of four. However, for better evaluation and interpretation of results, the researchers compared two different primers, SRP3 and SRP6, with the same crossbred sugarcane accessions to determine the common accessions to both primers that are Susceptible to the disease Smut. The results show that there are 16 offsprings that are highly likely to be susceptible to Sugarcane Smut: 216-1 (1), 216-4 (4), 216-6 (6), 216-8 (8), 218-3 (22), 218-4 (23), 218-10 (29), 219-3 (32), 219-4 (33), 219-5 (34), 219-6 (35), 219-9 (38), 220-1 (40), 220-2 (41), 221-4 (51), and 221-5 (52). According to the gathered and compared results, only the hybrid offspring 217, the cross between Phil 93-118-1207 (female, susceptible) and Phil 93-2349 (male, resistant), showed no susceptible samples. The rest of the hybrids show susceptibility. The specific hybrid offspring with the most number of susceptible samples is 219, the cross between Phil 2004-1011 (female, susceptible) and Phil 99-1793 (male, resistant), followed by 216, the cross between Phil 93-118-1207 (female, susceptible) and Phil 99-1793 (male, resistant). Moreover, when the accession Phil 99-1793 resistant is used as the male parent, some offspring show susceptibility no matter which female accession it is paired with. When Phil 93-118-1207, susceptible, is used as the female parent, the offsprings show no susceptibility, but when it is used as a male, some offsprings become susceptible. This study may be beneficial to the industry as the data gathered can be optimized for the creation of markers. This will quicken the pace of releasing varieties, hence improving the yield and quality of sugarcane in the industry.
Recommendations
From the results of the study, researchers recommend growing seedlings into the field and checking their phenotypic expression. Hypothetically speaking, they should be susceptible based on the garnered data. However, to confirm it, the varieties must be tested and observed in the field.
References
- Relles, J. M. G., Discaya, D. F. I., Mana-Ay, M. Z., Meneses, N. S., Entima, R. G., & Armones, R. T. Association Analysis of Developed Simple Sequence Repeat (SSR) Markers to Fungal Disease Resistance.
- Hasan, S. S., Solomon, S., Baitha, A., Singh, M. R., Sah, A. K., Kumar, R., & Shukla, S. K. (2015). CaneDES: A web-based expert system for disorder diagnosis in sugarcane. Sugar tech, 17(4), 418-427.
- Comstock, J. C. 2000. Smut. Pages 181-185 in: A Guide to Sugarcane Diseases. P. Rott, R. A. Bailey, J. C. Comstock, B. J. Croft, and A. S. Saumtally, eds. CIRAD and ISSCT, Montpellier, France.
- Rott, P., Bailey, R. A., Comstock, J. C., Croft, B. J., & Saumtally, A. S. (2000). A guide to sugarcane disease. CIRAD. ISSCT.
- Croft, B. J., Magarey, R. C., Allsopp, P. G., Cox, M. C., Willcox, T. G., Milford, B. J., & Wallis, E. S. (2008). Sugarcane smut in Queensland: arrival and emergency response. Australasian Plant Pathology, 37(1), 26-34.
- Bhuiyan, S. A., Croft, B. J., James, R. S., & Cox, M. C. (2012). Laboratory and field evaluation of fungicides for the management of sugarcane smut caused by Sporisorium scitamineum in seedcane. Australasian Plant Pathology, 41(6), 591-599.
- Que, Y., Xu, L., Wu, Q., Liu, Y., Ling, H., Liu, Y., Zhang, Y., Guo, J., Su, Y., Chen, J., Wang, S., and Zhang, W. (2014). Genome sequencing of Sporisorium scitamineum provides insights into the pathogenic mechanisms of sugarcane smut. BMC genomics, 15(1), 996.
- Sundar, A. R., Barnabas, E. L., Malathi, P., Viswanathan, R., Sundar, A. R., & Barnabas, E. L. (2012). A mini-review on smut disease of sugarcane caused by Sporisorium scitamineum. Botany, 2014, 226.
- Hasan, S. S., Solomon, S., Baitha, A., Singh, M. R., Sah, A. K., Kumar, R., & Shukla, S. K. (2015). CaneDES: A web-based expert system for disorder diagnosis in sugarcane. Sugar tech, 17(4), 418-427.
- Marques, Joao Paulo & Appezzato-da-Glória, Beatriz & Piepenbring, Meike & Massola, Nelson & Monteiro-Vitorello, Claudia & Vieira, Maria-Lucia. (2016). Sugarcane smut: Shedding light on the development of the whip-shaped sorus. Annals of Botany. 119. mcw169. 10.1093/aob/mcw169.
- Relles, J. M. G., Laurena, A. C., Diaz, M. G. Q., & Lalusin, A. G. (2018). Development of Microsatellite Markers From Sugarcane (Saccharum Officinarum) L.) Phil 97-3933. International Journal of Agriculture, Environment, and Bioresearch.
- Bailey, R. A. (1983). The effect of soil and seedcane applications of Triadimefon on the incidence of sugarcane smut (Ustilago scitaminea). In Proc S Afr Sug Technol Ass (Vol. 57, pp. 99-104).
- Bhuiyan, S. A., Croft, B. J., & Tucker, G. R. (2015). New method of controlling sugarcane smut using flutriafol fungicide. Plant disease, 99(10), 1367-1373.
- Simmons, M. (2017, November 14). Triazole — toxicity, side effects, diseases and environmental impacts. Retrieved from https://pesticides.news/2017-11-14triazole-toxici ty-side-effects-diseases-and-environmental-impa cts.html.
- Sharon, D., Adato, A., Mhameed, S., Lavi, U., Hillel, J., Gomolka, M., ... & Epplen, J. T. (1995). DNA fingerprints in plants using simple-sequence repeat and minisatellite probes. HortScience, 30(1), 109-112.
- Hameed, U., Pan, Y. B., Muhammad, K., Afghan, S., & Iqbal, J. (2012). Use of simple sequence repeat markers for DNA fingerprinting and diversity analysis of sugarcane (Saccharum spp) cultivars resistant and susceptible to red rot.
- Levi, A., & Rowland, L. J. (1997). Identifying blueberry cultivars and evaluating their genetic relationships using randomly amplified polymorphic DNA (RAPD) and simple sequence repeat-(SSR-) anchored primers. Journal of the American Society for Horticultural Science, 122(1), 74-78.
- Varshney, R. K., Graner, A., & Sorrells, M. E. (2005). Genic microsatellite markers in plants: features and applications. TRENDS in Biotechnology, 23(1), 48-55.
- Marín-García, J. (2011). Post-genomic cardiology. Academic Press.
- Kalia, R. K., Rai, M. K., Kalia, S., Singh, R., & Dhawan, A. K. (2011). Microsatellite markers: an overview of the recent progress in plants. Euphytica, 177(3), 309-334.
- Capra, M., Nuciforo, P. G., Confalonieri, S., Quarto, M., Bianchi, M., Nebuloni, M., ... & Draetta, G. F. (2006). Frequent alterations in the expression of serine/threonine kinases in human cancers. Cancer research, 66(16), 8147-8154.
- Garibyan, L., & Avashia, N. (2013). Research techniques made simple: polymerase chain reaction (PCR). The Journal of investigative dermatology, 133(3), e6.
- Bischoff, K. P., & Gravois, K. A. (2004). The development of new sugarcane varieties at the LSU AgCenter. J. Am. Soc. Sugar Cane Technol, 24, 142-164.